The Schneitz Lab - Shaping Beauty
How do organs know how big they have to grow? What controls the number, size and shape of organs? How does the cell wall affect those processes? These are the fundamental questions our lab is interested in. Tissue morphogenesis in plants depends on cell-cell communication as cells and groups of cells divide and expand in a coordinated fashion. A plant-specific complication relates to the fact that plant cells are encased by a semi-rigid extra-cellular matrix, the cell wall. The cell wall constitutes a barrier to the communication between cells. Moreover, it tightly connects plant cells to their neighbors. As a consequence plant cells do not migrate or intercalate and therefore tissue morphogenesis in plants depends on oriented cell division and unequal growth. It follows that plant cells have to constantly coordinate their behavior relative to their neighbors. In addition, the rigid coupling of cells brings about physical constraints and as a consequence mechanical stresses that contribute to morphogenesis. The molecular mechanisms underlying these complex signaling processes are poorly understood.
Signal Transduction at the Plasma Membrane - Cell Wall Interface


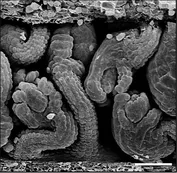
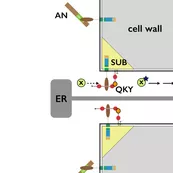
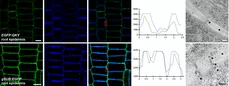
During evolution the obstacle of a cell wall has been overcome by at least two types of basic chemical signaling mechanisms. One involves trafficing of molecules through plasmodesmata (PD), membrane-lined channels interconnecting most plant cells. Another includes cell wall permeable ligands and their cellular receptors, for example cell-surface located receptor kinases. Our work on signal transduction mediated by the atypical receptor kinase STRUBBELIG (SUB) has revealed some of the first evidence for a functional connection between these two types of signaling processes.
Previously we could establish that signaling mediated by the atypical receptor kinase STRUBBELIG (SUB) is required for tissue morphogenesis in Arabidopsis (Chevalier et.al. 2005; Yadav et.al. 2008, Vaddepalli et.al. 2011). With QUIRKY (QKY), ZERZAUST (ZET) and ANGUSTIFOLIA (AN) we genetically identified three additional components of the SUB mechanism (Fulton et.al. 2009, Bai et.al. 2013). Presently, we are involved in resolving how exactly these factors interact during SUB signaling.
We could provide evidence that SUB and QKY function upstream of an unknown SUB-dependent mobile signal (SMS). Moreover, our data revealed that SUB and QKY physically interact at plasmodesmata (PD) providing evidence for functional crosstalk between receptor kinase- and PD-mediated signal transduction (Vaddepalli et.al. 2014). Thus, we are exploring the exciting hypothesis that SUB signal transduction controls movement of the SMS factor through PD.
Our work also revealed an exciting link between SUB signaling and the cell wall. ZET encodes an atypical β-1,3 glucanase (BG) that localizes to the cell wall (Vaddepalli et.al. 2017). Typically BGs degrade the cell wall polymer callose. Interestingly, however, ZET appears to be an atypical BG as our collective evidence indicated that its enzymatic activity is not required for function. Regardless, genetic evidence showed that ZET, SUB and QKY are all required for cell wall organization. It is now very interesting to investigate how SUB signaling controls tissue morphogenesis by connecting receptor-kinase-mediated signaling with cell wall and PD biology.
Extending the link of SUB signaling to cell wall biology we recently discovered a novel role for SUB in the control of cell wall integrity. In particular, SUB was found to play an important role in the cell's response to reduced cellulose in its cell wall (Chaudhary et.al. 2020). In particular, SUB is required for the compensatory cell wall remodelling that is induced by a reduction in cellulose content. We are presently exploring this exciting new function of SUB in the control of cell wall integrity.
Analysis of Ovule Curvature Using 3D Digital Ovules
A fascinating and fundamental problem relates to how morphogenesis integrates multiple processes acting at different scales, ranging from the molecular to the cellular and tissue levels. We use the ovule of Arabidopsis thaliana as a model system to study tissue morphogenesis. Ovules are the female reproductive organs of higher plants and carry the egg cell. Arabidopsis ovules (and ovules of many other species) are highly curved (anatropy). This curvature represents a unique and highly interesting aspect of morphogenesis. It is likely that an interplay of genetics, cellular coordination, and mechanical stresses due to the physical coupling of the cells controls this fascinating process. We would like to understand the molecular and cellular mechanisms regulating ovule curvature.
Ovules become apparent as finger-like protrusions that emanate from the placental tissue of the carpel (Schneitz et al., 1995; Robinson-Beers et al., 1992). Following primordium formation three proximal-distal (PD) pattern elements can be recognized. The distal nucellus, eventually harboring the embryo sac with the egg cell, the central chalaza which originates the integuments, and the proximal funiculus, a stalk-like structure connecting the ovule to the placenta. The two sheet-like integuments are determinate lateral organs of epidermal origin that undergo planar or laminar growth. Until shortly before fertilization each of the two integuments forms a bi-layered structure of regularly arranged cells. The inner integument initiates in a ring-like fashion while the outer integument initiates at the posterior side of the primordium but rapidly extends towards the anterior side. The integuments grow around the nucellus in an asymmetric fashion eventually forming a hood-like structure and contributing to the curved shape of the mature ovule.
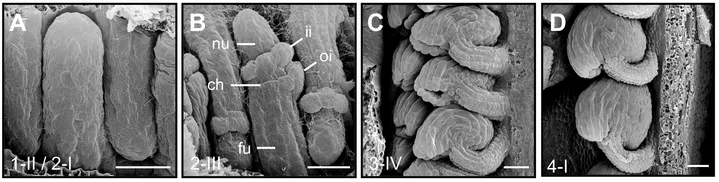
To address ovule morphogenesis, it is important to gain a quantitative understanding of the cellular growth patterns that generate the shape of the ovule. Accurate 3D digital ovules with cellular and tissue resolution are of central importance for this undertaking. The generation of 3D digital ovules requires deep imaging of ovules with reliable cellular resolution. Thus, we first established the necessary technology. As live imaging of entire ovules is currently not feasible the method is based on optically clearing fixed tissue with ClearSee (Kurihara et al., 2015), staining the cell contours with the cell wall stain SR2200 (Renaissance) (Musielak et al., 2015), and the nuclei with the fluorescent stain TO-PRO-3 iodide (TP3) (Van Hooijdonk et al., 1994). It is a fast and reliable method that allows deep imaging of entire Arabidopsis ovules in 3D and with cellular resolution using standard confocal microscopy. Importantly, it is compatible with a range of fluorescent proteins. A paper detailing the method has been published in PLANT METHODS (Tofanelli et al., 2019). In a collaboration with computer scientists we contributed to the development of the machine-learning-based PlantSeg pipeline for 3D cell boundary prediction and 3D cell segmentation (Wolny et al., 2020). We then used deep imaging of ovules coupled with the advanced image processing based on PlantSeg to generate 3D digital ovules with cell and tissue resolution (Vijayan, Tofanelli et al., 2021). To visualize and analyze 3D digital ovules we typically rely on the open source software MorphoGraphX (MGX) (Barbier de Reuille et al., 2015).
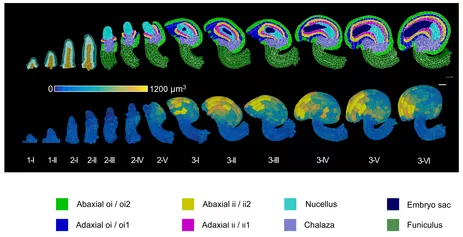
The left video reveals the two major PlantSeg steps: 3D cell boundary formation and 3D cell segmentation. After segmentation 3D cell mesh formation is done in MorphoGraphX. The right video shows a mature ovule. The heat maps indicates cell volume.
The video shows the 3D views that are possible once the major tissues have been annotated. Abbreviations: ii1, inner (adaxial) layer of inner integument; ii2, outer (abaxial) layer of inner integument; oi1, inner (adaxial) layer of outer integument; oi2, outer (abaxial) layer of outer integument.
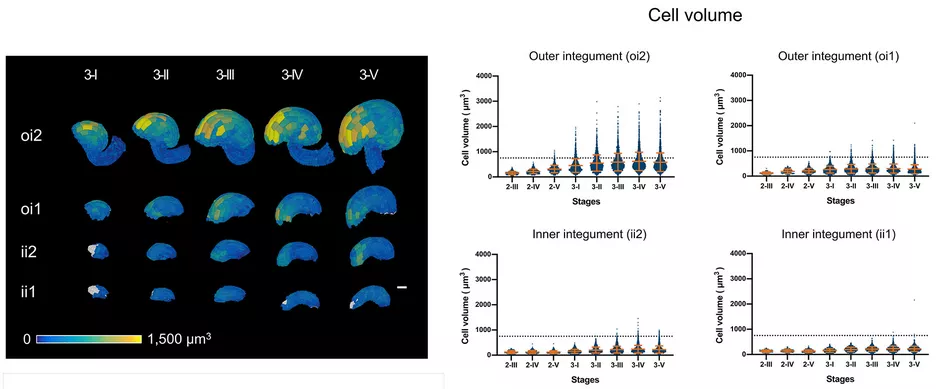
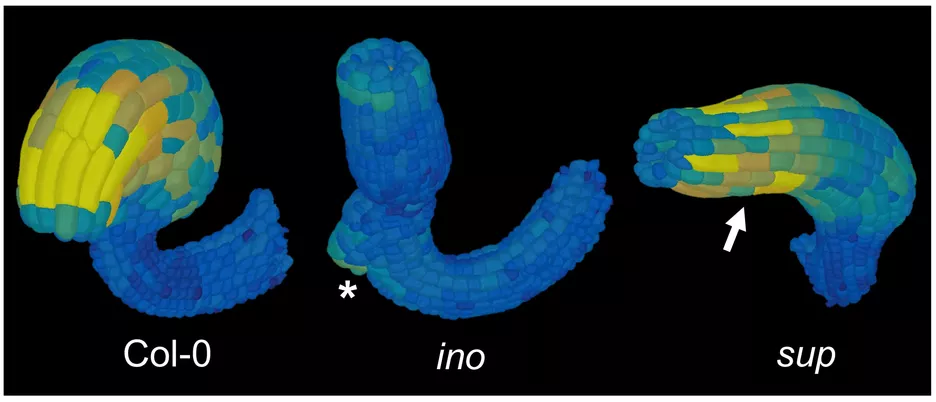
3D digital ovules allow a much more refined anatomical analysis of ovule development. For example, analysis of the cellular patterns of different tissues revealed novel insight into the cellular basis of ovule curvature. The results raise the possibility that curvature is in part caused by bending of the integuments. In particular, bending most likely involves differential enlargement and cell elongation in distal cells of the outer (oi2, abaxial) layer of the outer integument. Moreover, initial analysis of the inner no outer (ino) and superman (sup) mutants indicated that the outer integument is central to ovule curvature.
The quantitative analysis of ovule development is our contribution to the DFG-sponsored Research Unit "Plant Morphodynamics"